We have integrated novel experimental results with previous data about the roles of Nodal and BMP pathways in early development of Paracentrotus lividus in the form of a regulatory graph, which was complemented with logical rules, thereby enabling the simulation of cell responses to varying signalling inputs along the dorsal-ventral axis. This logical model was then extended to account for ligand diffusion and enable multicellular simulations, which accurately recapitulated gene expression in wild type embryos, accounting for the specification of the three main ectodermal regions, namely ventral ectoderm, ciliary band and dorsal ectoderm, and further recapitulated sophisticated mutant phenotypes. Finally, using a stochastic extension of the logical formalism, we performed more quantitative temporal simulations, which revealed a dominance of the BMP pathway over the Nodal pathway, and pointed to the rate of Smad activation as a key parameter for D/V patterning of the embryo.
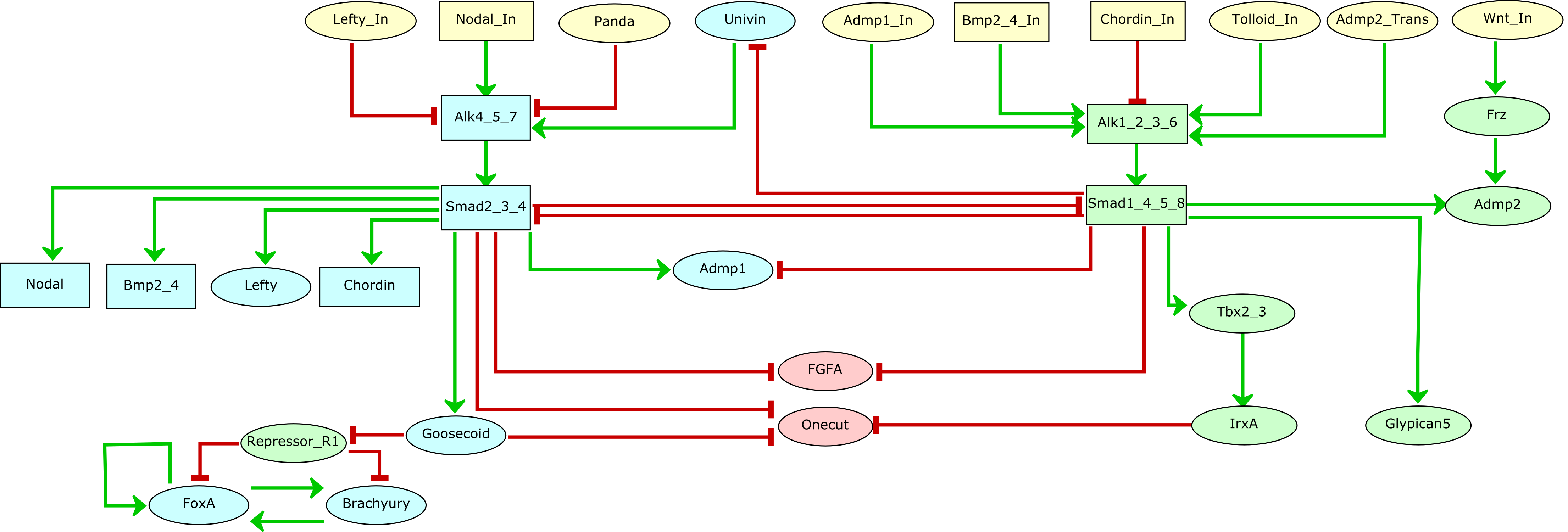
The links below enable the download of i) the unicellular GINsim model (zginml format, to be open with GINsim 3.0), the multicellular model (peps format, to be open with Epilog v1.1.1), and ii) the jupyter notebook containing the code of all the analyses (to be used with the colomoto-docker image).