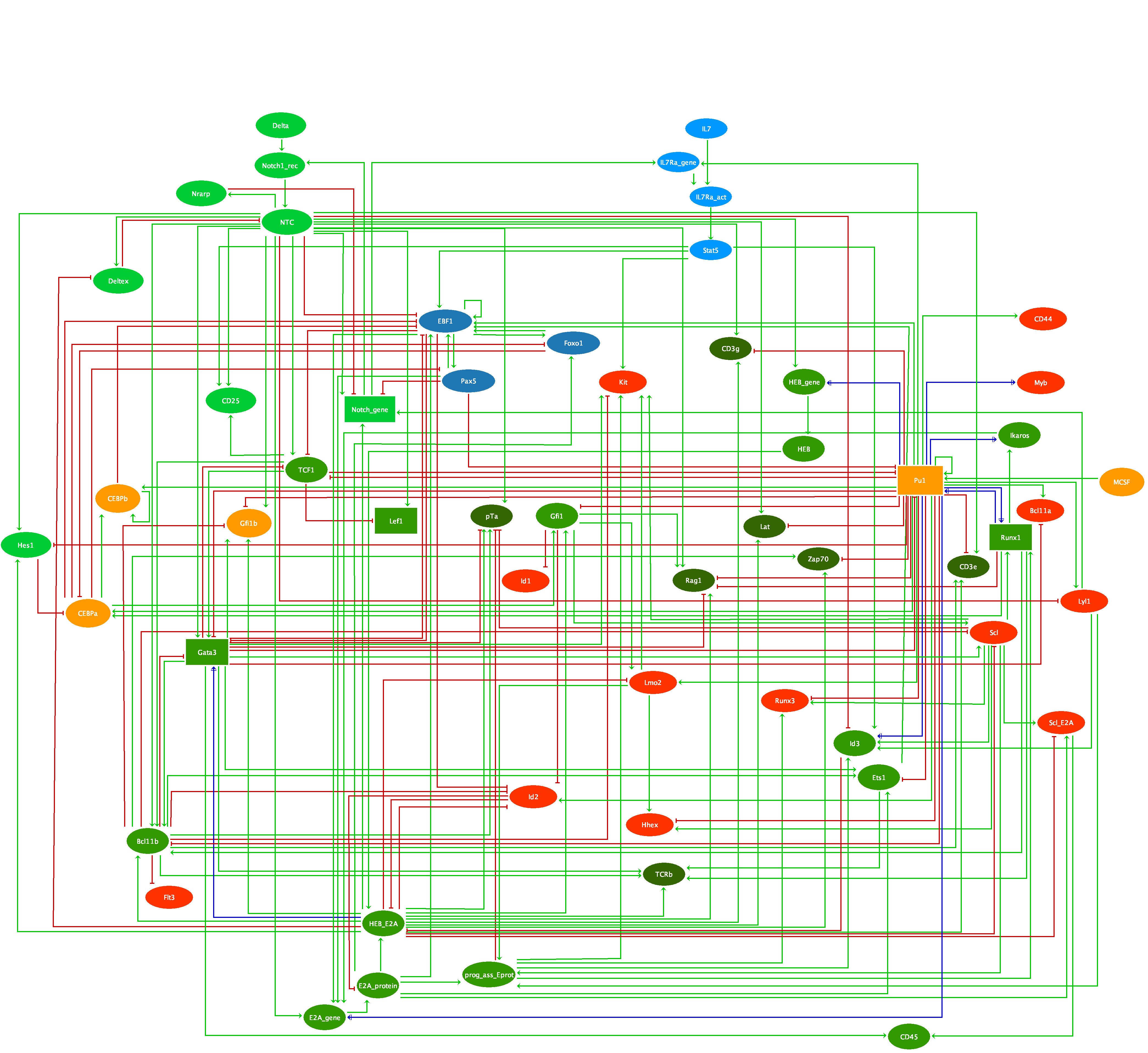
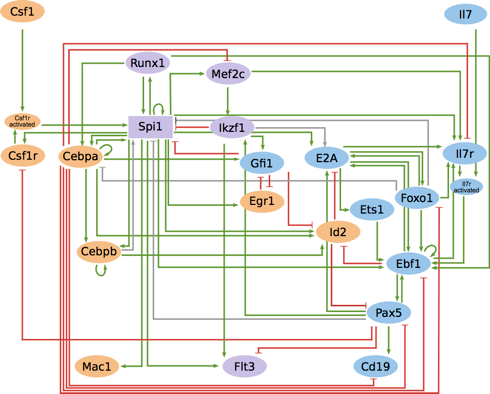
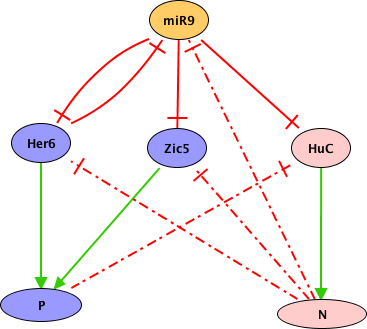
The vulva of Caenorhabditis elegans has been long used as an experimental model of cell differentiation and organogenesis. While it is known that the signaling cascades of Wnt, Ras/MAPK, and NOTCH interact to form a molecular network, there is no consensus regarding its precise topology and dynamical properties. We inferred the molecular network, and developed a multivalued synchronous discrete dynamic model to study its behavior. The model reproduces the patterns of activation reported for the following types of cell: vulval precursor, first fate, second fate, second fate with reversed polarity, third fate, and fusion fate. We simulated the fusion of cells, the determination of the first, second, and third fates, as well as the transition from the second to the first fate. We also used the model to simulate all possible single loss- and gain-of-function mutants, as well as some relevant double and triple mutants. Importantly, we associated most of these simulated mutants to multivulva, vulvaless, egg-laying defective, or defective polarity phenotypes. The model shows that it is necessary for RAL-1 to activate NOTCH signaling, since the repression of LIN-45 by RAL-1 would not suffice for a proper second fate determination in an environment lacking DSL ligands. We also found that the model requires the complex formed by LAG-1, LIN-12, and SEL-8 to inhibit the transcription of eff-1 in second fate cells. Our model is the largest reconstruction to date of the molecular network controlling the specification of vulval precursor cells and cell fusion control in C. elegans. According to our model, the process of fate determination in the vulval precursor cells is reversible, at least until either the cells fuse with the ventral hypoderm or divide, and therefore the cell fates must be maintained by the presence of extracellular signals.